Mitochondrial DNA Deletion Mutagenesis
Current Participant: Lakshmi Narayanan
While the mechanism of mitochondrial DNA (mtDNA) deletion mutagenesis is not precisely known, several hypotheses have been proposed, including the slip strand model and the DNA repair model (Figure 1). DNA misalignment involving direct repeats (DRs) is a prominent feature in these models, as many deletions have breakpoints (i.e. DNA positions adjacent to the DNA segment that is lost) that are either near or exactly flanked by DRs. However, the close proximity of deletion breakpoints to these sequence motifs can occur by random chance, especially considering that DR motifs can be readly found along human mtDNA sequence. Moreover, some of the observed breakpoints are not near any DR at all. Whether these motifs are actually involved in the deletion process and whether there is any species-specific role of such motifs, require a careful evaluation.
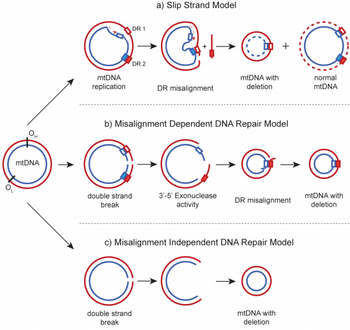
In our study, we have analyzed mitochondrial DNA deletion breakpoints and mitochondrial genomes from four different mammals: human, rhesus monkey, mouse and rat. Our analysis gave evidence for the causal role of DRs in the formation of age-related mtDNA deletions, only among the long-living mammals, i.e. human and monkey. A further analysis of the lifespans and mtDNA sequences of 294 mammalian and 236 avian species, the most comprehensive of such analysis to date, further showed that contrary to a long-held hypothesis, the number of DRs is not a specific, evolutionarily selected feature of mtDNA. Instead, DRs form as an indirect consequence of bias in nucleotide composition and in synonymous codon usage. Taken together, our findings reveal a lack of negative selection pressure against DR motifs in mtDNA, even among long-lived species, despite their role in mtDNA deletion formation.
References
- Lakshmanan, LN., Gruber, J., Halliwell, B. and Gunawan, R. Are mutagenic non D-loop direct repeat motifs in mitochondrial DNA under a negative selection pressure?. Nucleic Acids Research 43(8): 4098-4108 (2015). external pageabstractcall_made
- Lakshmanan, LN., Gruber, J., Halliwell, B. & Gunawan, R. Role of direct repeat and stem-loop motifs in mtDNA deletions: cause or coincidence? PLoS One 7, e35271 (2012). external pageabstractcall_made
